Coherent Diffractive Imaging of Single Particles
Our group develops novel methods to achieve the foundational goals of single-particle diffractive imaging using XFEL pulses.
Single-particle diffractive imaging is one of the key foundational goals behind the establishment of X-ray free-electron laser (XFELs) facilities and has the potential to revolutionize the way single-molecules are imaged. Single-molecule diffractive imaging at ultrafast time-scales of atomic and electronic motions is expected to provide a wealth of information to map the complete conformational landscape of molecules both at ground and excited states, without the need for cryogenic cooling and which otherwise can not be obtained through conventional molecular imaging methods [1].
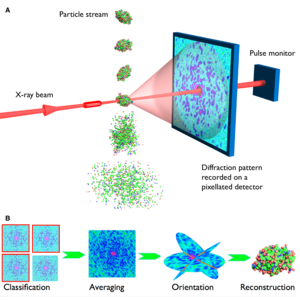
Extremely intense femtosecond XFEL pulses outrun radiation damage and open up the possibility of radiation-damage-free high-resolution imaging of nanoparticles, macromolecules, and viruses at room temperature. Although XFELs impart large doses onto matter in a single pulse, the short femtosecond pulse duration plays the key role in outrunning radiation damage effects [2]. The extreme intensity—a billion times brighter than beams produced at synchrotron radiation facilities—provide enough photons per pulse to obtain measurable signals from small objects such as uncrystallized aperiodic single-particles. This may obviate the need for crystals—overcoming the need of crystallizing macromolecules. As in crystallography, the spatial coherence of the incident XFEL pulse leads to a diffraction pattern (recorded on a detector in the far field) that represents the two-dimensional Fourier transform snapshot of the electron density of the sample. However, without the repetition of the crystal lattice, this diffraction pattern does not consist of a set of isolated Bragg peaks but is continuously varying. This pattern exhibits speckles or undulations of a size that are inversely proportional to the extent of the object. As in crystallography, many such patterns are measured—here from a supply of identical objects—and combined to build up a Fourier representation of the object of interest. These Fourier amplitudes must then be used to synthesize the actual real-space image of the object, but this can only be done by first assigning phases to the measured amplitudes. In crystallography, that step usually requires additional information, in the form of chemical restraints of the molecular structure, for example, or other diffraction datasets collected at wavelengths that straddle atomic resonances. But with the complete continuous molecular transform available in single-particle imaging, iterative phasing algorithms can recover the phases directly to solve the electron density map of the target structure (see Figure 1). Compared to the real-space imaging with lenses, diffractive imaging is aberration free, and may be suitable for investigating irreversible or non-cyclic processes at femtosecond timescales [3].
The XFEL pulses incident on the target particles must be short enough to carry the information or the diffraction pattern of the target object in the scattered photons before the particle is destroyed due to the onset of radiation damage processes initiated by photo-ionization, which lead to displacements of atoms, a change in atomic scattering properties, and an eventual Coulomb explosion of the object. This process of “diffraction-before-destruction” allows the pristine radiation-damage-free state of the target to be recorded. Since only one two-dimensional snapshot can be taken per pulse, a continuous stream of reproducible particles needs to be supplied in a serial-fashion to record enough noisy 2D single-shots for the reconstruction of a complete 3D structure (see Figure 1) [3]. The number of required shots depends on the size of the target, the signal-to-noise ratio, and the desired resolution. We helped demonstrate the principle of “diffraction-before-destruction” using the world's first free-electron laser FLASH in 2006 and showed that coherent diffractive imaging (CDI) of non-periodic objects can be successfully performed with femtosecond FEL pulses [4]. Subsequently, we demonstrated femtosecond time-delay X-ray holography [5], nanoscale ultrafast dynamics of exploding 2D objects at FLASH [6], and the first single-particle diffractive imaging of free-flying aerosolized nanoscale objects with FELs at FLASH in 2007 [7]. Nanoparticles of a sucrose-DNA mixture were electrospray injected, aerosolized, and delivered to the FEL beam using an aerodynamic lens stack [7]. And subsequently, in 2008, we introduced the method of holographic-reference-based diffractive imaging of weakly scattering single-particles [12].
Since the first proof-of-principle experiments carried out with collaborators from Lawrence Livermore National Laboratory, Uppsala University, SLAC, Arizona State University and elsewhere, we have continued to pursue the development of single-particle imaging (SPI) experiments at XFEL facilities together with our collaborators, for example in imaging giant viruses [8, 9] and aerosol soot nanoparticles [10]. We also contribute to the community-led SPI initiatives at the Linac Coherent Light Source (LCLS), Stanford, USA [11] and the European XFEL (EuXFEL), Hamburg, Germany, and develop SPI experiments and techniques.
Figure 1. XFEL single-particle diffractive imaging pipeline involves recording single-shot diffraction patterns of the target object before its destruction due to ionization and Coloumb explosion (top), computational purification of single-particle diffraction patterns, orientation determination, classification into 2D class averages, and the 3D merging of intensities in the reciprocal space. The Fourier magnitudes are then used to synthesize a real-space image of the object, but only after recovering the phase information that is missing from the diffraction experiment. Phasing is achieved using iterative phase retrieval algorithms (bottom). Image source: Henry Chapman, CFEL. Science, 2007, 316, 1444-48.
The challenge: Since single macromolecules or even whole viruses scatter very weakly, achieving high-resolution 3D reconstruction of a macromolecule has been a longstanding challenge, and requires developments in the direction of sample delivery methods—friendly to native or physiological states of macromolecules—providing a high fraction of diffraction patterns, while simultaneously achieving low-background scattering, novel x-ray optics, and sensitive detectors with high dynamic range. Meanwhile, in the voyage of imaging single molecules, we demonstrated the benefit of XFELs by developing the popular serial femtosecond crystallography method using tiny crystals to image biologically important macromolecules, enzyme kinetics and light-induced ultrafast dynamics at room temperature [1].
We investigate and develop many aspects required for high-resolution single-particle imaging, such as instrumentation, x-ray optics, holographic methods, sample preparation and characterization, low-background sample delivery methods, online and offline data-analysis, image classification and reconstruction algorithms, and software. We are interested in applying these development to the structure determination of inorganic nanoparticles, biological macromolecules, fibers, viruses, and artificial nucleic acid nanostructures. As an innovative approach, the group also focuses on amplifying weak single-molecule signal through arbitrary alignment of macromolecules and deploying strongly scattering holographic references to enhance the signal of weakly diffracting single-molecules [12].
Though sub-nm resolution single-particle x-ray scattering signal has been observed with large viruses [1], the current state-of-the-art three-dimensional reconstruction of a virus or any bio-particle is around 6.9-nm resolution (as of 2022) and sub-3 nm resolution in case of metallic nanoparticles, which we have demonstrated recently by collecting more than ten-million single-particle diffraction patterns of anisotropic AuNPs at the EuXFEL [13]. Achieving high-resolution single-macromolecular structure determination with x-ray lasers is a challenge which requires multidisciplinary efforts and developments [14,15]. With the advent of high-repetition-rate megahertz XFELs, improved x-ray optics, innovative holographic, sample-preparation and delivery methods unique to XFEL imaging conditions, and efficient algorithms coupled with deep learning, soon we are set to enter the domain of sub-nm resolution, million-pattern XFEL imaging of single-particles.
DNA NANOTECHNOLOGY
Acknowledgements
Human Frontier Science Program (HFSP RGP0010/2017), European Research Council - Frontiers in Attosecond X-ray Science: Imaging and Spectroscopy (ERC-AXSIS), CUI: Advanced Imaging of the Matter, DESY-Helmholtz Association